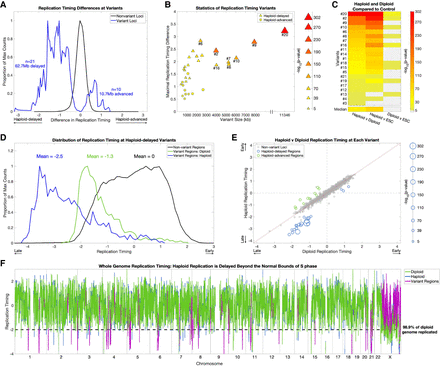
Replication timing in haploid cells is delayed beyond the normal S-phase. (A) Haploid-delays are the predominant form of replication timing variation. Distributions of the replication timing differences between haploid and diploid cells (mean of each pair of cell lines) in nonvariant and variant regions. The distributions represent individual windows (2.5 kb) in each category. (B) The size (scaled by the −log10[P-value]), absolute replication timing difference, and variation P-value of all 31 haploid-diploid replication timing variants. Haploid-delayed variants segregate from haploid-advanced variants on all three metrics, shown by the clustering of haploid-advanced (circles) in the bottom left, as well as their weaker significance. Conversely, haploid-delayed variants (triangles) are longer, have a greater replication timing difference from diploids, and are more significant. (C) Replication timing variation is attributed to alterations in haploid cells. One-way ANOVA P-values between haploid, diploid, and control ESC lines (CU-ES4 and CU-ES5, shown as “ESC”) at haploid-delayed variants, sorted by P-value of haploid versus diploid comparisons (leftmost column). Light gray: not significant. Nineteen of the 21 haploid-delayed variants are also significantly different from controls (middle column), in contrast to only nine variants that vary between diploid cells and ESCs (rightmost column). In all but one case (variant #13), haploid-ESC variation is more significant than diploid-ESC variation, thus haploids, not diploids, have the exceptional replication timing values. (D) Replication timing variants are normally late-replicating regions that are further and severely delayed in haploids. Distributions of replication timing for diploid cells in nonvariant regions (i.e., normal replication timing distribution; black), and for diploid (green) and haploid (blue) cells in haploid-delayed variant regions. The distributions represent individual windows (2.5 kb) in each category. Haploid-delayed variants replicate late (on average, 1.3 SD later than the genome mean) in diploid cells and considerably later (2.5 SD later than the mean and beyond the bounds of normal S phase) in haploid cells. (E) The majority of haploid-delayed variants are already late-replicating in diploid cells. Scatterplot of haploid versus diploid replication timing across the genome. Nonvariant loci were defined by first removing variants, then binning the genome into 2.3-Mb regions (mean size of all replication timing variants). Haploid-delayed variants (blue) replicate late in diploid cells and even later in haploids. The size of data points is scaled by −log10(P-value). The three dots (corresponding to variants #3, #4, and #13) with both early haploid and diploid replication had the poorest P-values of all haploid-delayed variants (P = 7.9 × 10−6, 2.3 × 10−10, and 2.8 × 10−10). (F) Replication in haploid cells is delayed beyond the bounds of S-phase. The dashed line indicates a replication timing of 2 SDs below the mean, when 98.9% of the genome has already completed replication. Eighteen of the 21 replication timing delays (all but the three least significant variants #3, #4, and #13) extended beyond this value.











