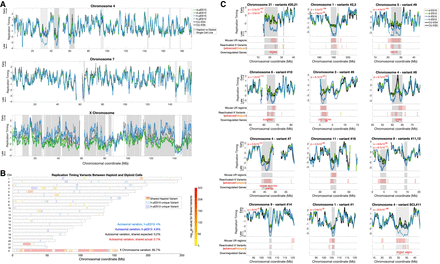
Significant replication timing variation between haploid and diploid ESCs. (A) Representative replication timing profiles for haploid and diploid pES10 and pES12 and control ESCs. Profiles are z-score-normalized to a genome-wide average of zero and a standard deviation of one, such that positive and negative values indicate replication timing that is earlier and later than average, respectively, and the units correspond to standard deviations (SD). Gray: regions of variation between haploid and diploid cells (light: one cell line; dark: shared in both cell lines). (B) A genome-wide view of all replication timing variants. Shared variants are color-coded by P-value. The degree of shared haploid variation is significantly greater than expected given the extent of single cell line variation (χ2 test P << 10−16). (C) The 11 most significant haploid-delayed variants, numbered by genomic location and ordered by P-value. Variants #21, #3, and #12 are each proximal to another, larger variant (represented by the second-listed P-value in each panel). Variant SCL#11 (single cell line variant #11) is only delayed in a h-pES12 but nonetheless shows features common to other haploid-delayed variants (see text). Shown below each variant are the locations of all mouse UR regions (those found in all stages of development) and reactivated-X variants in the plotted interval of each panel. Reactivated-X delays are shown in red, and advanced regions are in orange. Down-regulated genes are shown only within the replication timing variant borders (gray shades). Supplemental Figure S3 shows all remaining variants.











