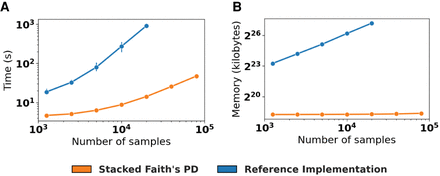
SFPhD outperforms the reference implementation in terms of runtime and memory usage. (A) Runtime in seconds for computing Faith's PD on data sets with thousands of samples and 100,000 tips in the phylogeny. Data are independently subsampled from a collection of 113,721 public samples in Qiita (Gonzalez et al. 2018; Zhu et al. 2019) as previously processed (McDonald et al. 2018b). Mean of n = 10 repetitions with 95% CI error bars. (B) Memory usage for the same experiment as in A. For both A and B, jobs were terminated if they exceeded 250 GB of memory.











