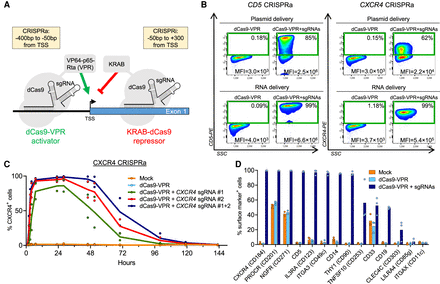
CRISPRa by plasmid and RNA-based delivery. (A) CRISPRa or CRISPRi is mediated by a nuclease-deactivated Cas9 (dCas9) fused to effector domains that either activate (VP64-p65-Rta [VPR]) or inhibit (KRAB) transcription. The fusion protein is guided by one or more sgRNAs targeted to the region surrounding the transcriptional start site (TSS). The guidelines for sgRNA positioning relative to the TSS have been empirically devised by Gilbert et al. (2014) and are shown in the yellow boxes. (B) Representative FACS plots (N = 3) showing the analyses of target gene expression in K562 cells 24 h after electroporation with CRISPRa systems based on either plasmid or RNA delivery. For the plasmid-based platform, one plasmid encodes dCas9-VPR, and separate plasmids each express a sgRNA targeting the TSS region of the target gene (CD5 or CXCR4). The RNA-based platform is based on in vitro transcribed mRNA encoding dCas9-VPR and synthetic, chemically modified sgRNAs. Four sgRNAs were used for CD5, and two sgRNAs were used for CXCR4. Gates contain cells that are positive for the target protein with percentages shown in the upper right corner. Each FACS plot also displays the mean fluorescence intensity (MFI) of all live cells. (C) Time course experiment showing the percentage of CXCR4+ cells measured by flow cytometry following electroporation with dCas9-VPR mRNA and two chemically modified sgRNAs. (D) CRISPRa of 14 different target genes in K562 cells. Cells were analyzed by flow cytometry 24 h after electroporation with dCas9-VPR mRNA and chemically modified sgRNAs (two to four sgRNAs per gene). The percentage of cells positive for the surface marker is shown. For all graphs, N is number of data points, and all bars show mean values with individual data points plotted.











