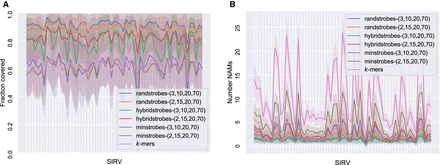
Comparison between strobemers and k-mers when matching ONT cDNA reads (7.0% median error rate) to 61 unique spike-in RNA variant (SIRV) reference sequences. Each SIRV corresponds to a tick on the x-axis. (A) Total fraction of the SIRV covered by NAMs from reads (y-axis). (B) The number of NAMs (y-axis) between a read and the SIRV. The line shows the mean, and the shaded area displays the standard deviation of the reads. A high NAM coverage and low number of NAMs means long contiguous matches and facilitates accurate and efficient sequence comparison.











