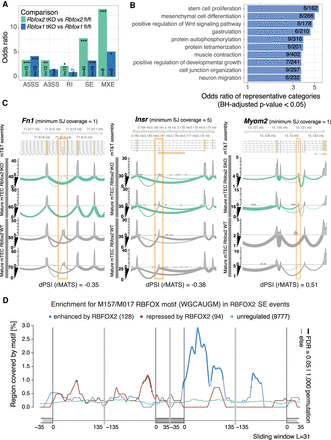
RBFOX regulates alternative splicing in TEC. (A) The bar plots show the enrichments (ORs) of previously predicted Rbfox target genes (Weyn-Vanhentenryck et al. 2014) in the sets of genes differentially spliced in the Rbfox1 tKO or Rbfox2 tKO mature mTEC (two-sided Fisher's exact tests, BH adjusted P-values): (*) P < 0.05, (**) P < 0.01, (***) P < 0.001; abbreviations as for Figure 4B. (B) Selected GO biological processes significantly overrepresented (one-sided Fisher's exact tests, BH adjusted P-values < 0.05) in genes that contained significantly RBFOX2-regulated SE events in mature mTEC. (C) Three examples of significantly (FDR < 0.05) RBFOX2-regulated splicing events in mature mTEC. Fn1 and Insr are known RBFOX target genes (Chen and Manley 2009; Weyn-Vanhentenryck et al. 2014). Myom2 is an example of a non-Aire TRA gene. (D) Enrichment of the RBFOX recognition motif (M159/M017) (Ray et al. 2013) in the sequences surrounding exons that were found to be significantly regulated by RBFOX2 in mature mTEC (Supplemental Fig. 18B). The lines show enrichments for sets of exons found to be enhanced (blue), repressed (red), or not significantly regulated (green) by RBFOX2. Thicker lines indicate regions of statistically significant enrichment (FDR ≤ 0.05, n = 1000 permutations).











