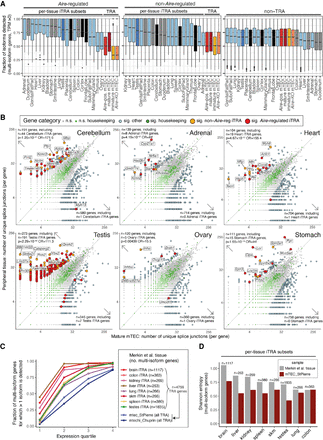
mTEC express fewer isoforms of TRA genes than peripheral tissues. (A) The fraction of isoforms detected (TPM > 0) per multi-isoform protein-coding gene in mTEC and ENCODE peripheral tissues. The box plots show the fractions for Aire-regulated TRA genes (left), non-Aire TRA genes (middle), and non-TRA genes (right, colors as in Fig. 1) (Supplemental Fig. 3D). (B) The numbers of splice junctions found in protein-coding genes (points) in mature mTEC (this study; x-axes) versus peripheral tissues (ENCODE Project; y-axes). Six selected peripheral tissues are shown, with the remaining 14 displayed in Supplemental Figure 5A. Significant (sig.) differences in junction number were identified using edgeR (BH adjusted P < 0.05, |fc| > 2). P-values and odds ratios (OR) from Fisher's exact tests for enrichment of iTRA genes among the genes with significantly higher junction counts in the peripheral tissues are reported (top left). The top 10 of each tissue's iTRA genes with significant differences in junction counts are labeled (as ranked by edgeR P-value). (C,D) Validation with independent peripheral tissue (Merkin et al. 2012) and TEC data sets (St-Pierre et al. 2013; Chuprin et al. 2015). (C) The fraction (y-axis) of alternatively spliced multi-isoform genes (>1 splice isoform detected) is shown for individual sets of tissue iTRA genes by expression quartile (x-axis). (D) The bar plots show the mean Shannon entropy (y-axis) of splice isoform expression for sets of multi-isoform tissue iTRA genes (x-axis). Alternate versions of C and D made using all genes are shown in Supplemental Figure 5B and 5C. For A and C, the full sets of TRA genes were quantitated in the TEC populations.











