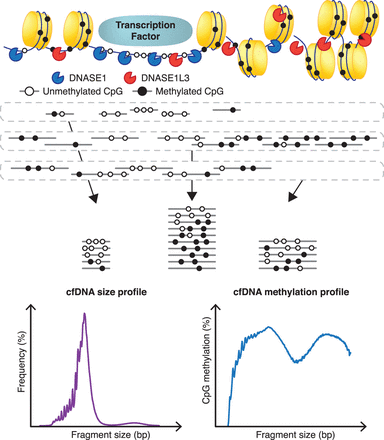
Deduced activities of DNASE1 and DNASE1L3. DNASE1 (blue) prefers to cleave unmethylated and open chromatin DNA. By fragmenting these regions, DNASE1 increases the representation of these OCR and CGI regions in plasma, especially in fragments sizes ≤80 bp, resulting in the relative hypomethylation of cfDNA. These OCR and CGI regions are unequally represented among different cfDNA sizes. DNASE1L3 (red) is effective at cutting methylated fragments and increases the representation of methylated fragments in plasma cfDNA compared with DNASE1. DNASE1L3's cutting preference likely results in the prominence of the 166-bp fragment size. The combination of these preferences leads to the eventual cfDNA size profile and methylation profile observed for each fragment size.











