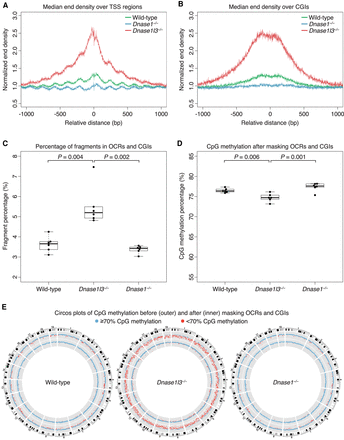
OCR and CGI fragments have a prominent role in the relative hypomethylation of cfDNA. (A,B) The normalized end density is calculated from fragment end counts divided by the median end counts in the ±3000 bp region. The median normalized end density for each genotype is shown in a ±1000-bp window over the aggregated TSS region (A), and CGI regions (B). cfDNA from wild-type mice is in green, DNASE1L3-deficient mice is in red, and DNASE1-deficient mice in blue. The fragmentation of cfDNA from DNASE1L3-deficient mice is increased and decreased in DNASE1-deficient mice. The y-axis scale ranges from 0.5% to 3%. (C) The regions ±500 bp around the center of TSS, PoI II, H3K4me3, and H3K27ac regions were merged with CGI regions. The proportion of fragments in these OCR and CGI regions are shown. cfDNA from DNASE1L3-deficient mice had a significantly increased proportion of OCR and CGI fragments. Welch's t-test was performed for significance testing. The y-axis scale ranges from 2% to 8%. (D) OCR and CGI fragments were bioinformatically excluded in masking analysis. The CpG methylation percentage increased after these fragments were masked. The relative hypomethylation of cfDNA from DNASE1L3-deficient mice is substantially diminished after masking but remains significantly different from the cfDNA methylation of wild-type mice. Welch's t-test was performed for significance testing. The y-axis scale ranges from 55%–85%. (E) Circos plots showing genome-wide CpG methylation percentages before (outer ring) and after (inner ring) masking OCR and CGI fragments. Each dot represents the CpG methylation percentage in a 1-Mb bin of the mouse autosome and is colored in blue if ≥70% and in red if <70%. Representative Circos plots are shown for wild-type, DNASE1L3-deficient, and DNASE1-deficient mice.











