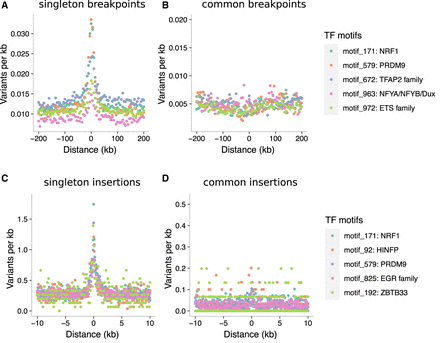
Binding factors associated with the highest rates of mutation at spermatogonial binding sites. Plots are centered on the binding sites of a given motif family inside ATAC-seq footprints. (A,B) Singleton (A) and common (B) deletion breakpoints in the DDD cohort; singletons are breakpoints of deletions with a frequency of ∼0.002% across population samples; common variants are seen at a frequency of at least 1% in the DDD consensus data set. (C,D) Singleton (C) and common (D) insertions (5–20 bp) in the gnomAD data set. Singletons are seen only once in gnomAD V.3 (allele frequency ≤0.001%), and common variants have an allele frequency of at least 5% within gnomAD V.3. Only 10-kb regions around TFBSs with ≥95% unique mappability (umap24 scores) were included. The top five disrupted motifs are shown, listed in order of enrichment of singleton variants in the circular permutations (all enrichments of singletons are associated with P-values < 10−4).











