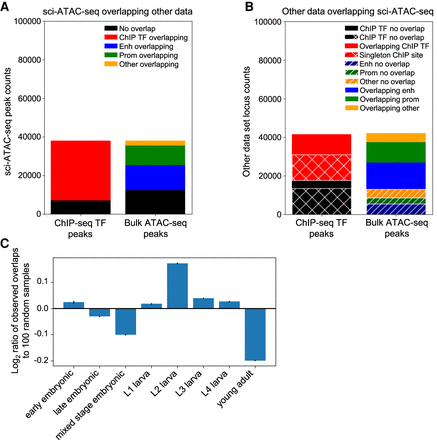
The peaks called from sci-ATAC-seq data show substantial overlap with existing chromatin data collected from whole worms. (A) The majority of sci-ATAC-seq peaks overlap sites called in the transcription factor (TF) ChIP-seq peaks from modERN or the bulk ATAC-seq peaks from Jänes et al. (2018). (B) Peaks from the other data sets also show substantial overlap with sci-ATAC-seq peaks. Most of the ChIP-seq TF peaks that do not overlap a sci-ATAC-seq peak are singleton peaks that are only found in a single experiment. (C) Breaking out the ChIP-seq peak overlaps by the developmental stage of the worms assayed and comparing the distribution across stages of the peaks with overlaps compared with the stage distribution for randomly selected ChIP-seq peaks show an enrichment for peaks found in larval stage L2. Error bars, 95% confidence interval.











