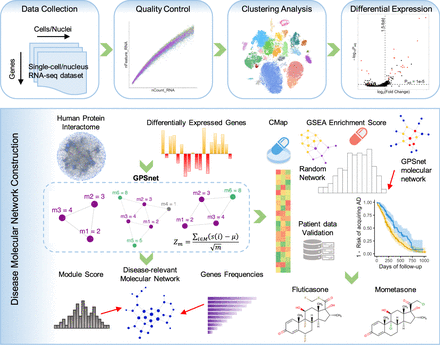
A diagram illustrating the network-based framework. A standard single-cell/nucleus RNA sequencing (sc/snRNA-seq) data analysis pipeline includes quality control, clustering analysis, and differentially expressed gene (DEG) analysis. We built the molecular network using the state-of-the-art network-based algorithm (termed GPSnet) by integrating sc/snRNA-seq data into the human protein–protein interactome (Methods). Next, we prioritized repurposed drugs for potential treatment of Alzheimer's disease (AD) by identifying those that specifically reverse dysregulated gene expression for molecular networks of disease-associated microglia (DAM) or astrocyte (DAA): if drug-induced up- or down-related genes are significantly enriched in the dysregulated molecular networks, these drugs will be prioritized as potential candidates for treatment of AD. Finally, top drug candidates were validated further using a large-scale, longitudinal patient database. (GSEA) Gene set enrichment analysis; (CMap) connectivity map.











