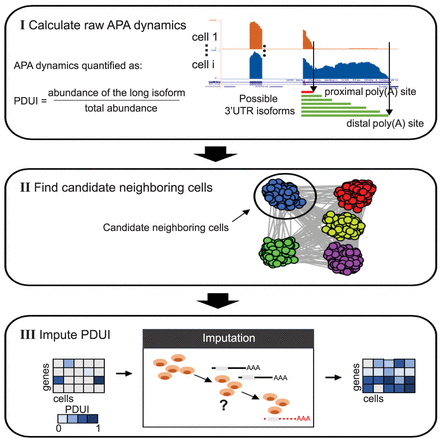
A schematic illustration of the scDaPars algorithm. (I) scDaPars predicts both distal and proximal poly(A) sites by joint analysis of all single-cell samples and quantifies the raw relative APA usage by the proportion of estimated abundances of transcripts with distal poly(A) sites (long isoform). (II) scDaPars determines potential neighboring cells by applying community detection methods in APA profiles generated in step I. (III) scDaPars uses the NNLS regression model to refine neighboring cells and impute missing values by borrowing APA information from neighboring cells.











