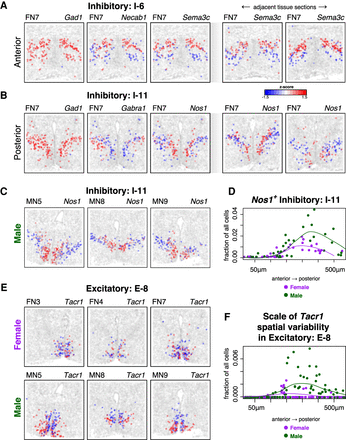
MERINGUE identifies spatial heterogeneity within cell types in the preoptic region of the mouse hypothalamus using MERFISH. (A, left) Expression of three sample genes in Inhibitory I-6 neurons in female naïve animal 7 (FN7). Each point is a cell. Cells are colored by expression with red denoting high expression and blue denoting low expression. Cells that are not I-6 cells are colored in gray. Gad1 is highly expressed in all I-6 neurons, whereas Necab1 and Sema3cshow significant spatial variation. (Right) Expression of Sema3c in I-6 in adjacent tissue sections in FN7 show similar spatial patterning. (B, left) Expression of three sample genes in Inhibitory I-11 neurons in FN7. Again, Gad1 is highly expressed in all I-11 cells, but Gabra1 and Nos1 show significant spatial variation. (Right) Expression of Nos1 in I-11 neurons in adjacent tissue sections in FN7 show similar spatial patterning. (C) Expression of Nos1 in I-11 neurons in representative male naïve animals MN5, MN8, and MN9 show similar spatial patterns to the female animal in B. (D) Fraction of Nos1+I-11 cells for male and female animals across tissue layers from the anterior to posterior preoptic region. Each dot represents one tissue layer in one animal. Lines represent fitted curves for males and female animals. (E) Expression of Tacr1 in E-8 neurons in female (top) and male (bottom) animals. (F) Scale spatial heterogeneity of Tacr1 in E-8 neurons for male and female animals across tissue layers from the anterior to posterior preoptic region. Each dot represents one tissue layer in one animal. Lines represent fitted curves for male and female animals.











