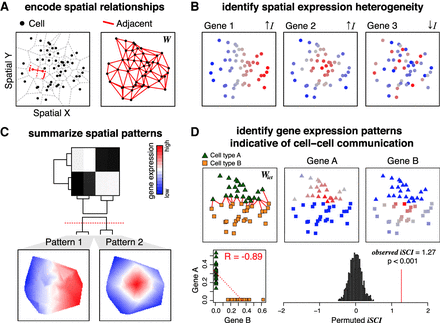
Overview of MERINGUE. (A) MERINGUE encodes spatial relationships among spatially resolved data sets, such as cells, using a binary adjacency weight matrix W. (Left) Two cells are considered adjacent if their neighborhoods inferred from Voronoi tessellation share an edge. (Right) The binary adjacency weight matrix W is visualized by plotting all cells in space with a red line connecting cells if cells are adjacent and no line otherwise. (B) MERINGUE identifies genes with spatially heterogeneous expression using W to compute and evaluate the significance of a spatial autocorrelation index I for each gene. When a gene's expression magnitude between spatially adjacent cells are highly correlated, I will be positive, indicative of spatial heterogeneity. Three simulated gene expression profiles are provided to illustrate examples of high and low spatial heterogeneity with red indicating high expression and blue indicating low expression. (C) MERINGUE groups identified spatially heterogeneous genes into primary spatial patterns by computing a spatial cross-correlation index for every gene pair. (Top) The resulting spatial cross-correlation matrix is used to construct a hierarchical dendrogram. (Bottom) Dynamic tree cutting is applied to partition genes into patterns. Groups of genes are Z-scored and averaged, with empty regions filled in using Akima interpolation to visualize final patterns. (D) MERINGUE identifies gene expression patterns that may be indicative of putative cell–cell communication using an inter-cell-type weight matrix Wict between two cell types, which can then be used to compute an inter-cell-type spatial cross-correlation index iSCI between two genes (Top left). Two cell types, A and B, are shown as green triangles and orange squares, respectively. Wict is visualized with a red line for cells of cell type A spatially adjacent to cells of cell type B. (Top right) Cell type–A cells express gene A at variable levels, whereas cell type–B cells express gene B at variable levels, with red indicating high expression and blue indicating low expression. (Bottom left) Cell type–A cells do not express gene B and cell type–B cells do not express gene A, resulting in a generally negative Pearson's correlation (R) between the two genes. (Bottom right) However, expression of gene A in cells of cell type A is highly correlated with the expression of gene B in spatially adjacent cells of cell type B, resulting in a positive iSCI. The significance of this iSCI is assessed by permutation.











