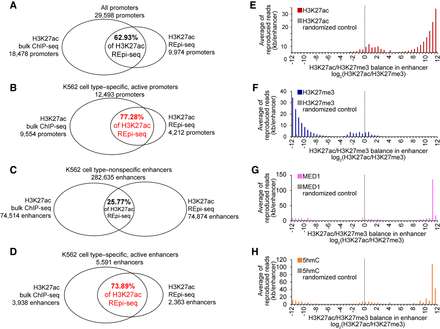
Evaluation of REpi-seq reproduced reads. (A–D) REpi-seq reproduced reads evaluated by bulk ChIP-seq. H3K27ac reproduced reads from eight single cells were combined before statistical analysis. Enrichment of H3K27ac reproduced reads was analyzed in all promoters (A), K562 cell type–specific, active promoters (B), K562 cell type–nonspecific, active enhancers (C), K562 cell type–specific, active enhancers (D). Significance of reproduced read enrichment in promoters and enhancers was calculated by the bootstrap procedure. Promoters and enhancers enriched with reproduced reads (false-positive rates <5%) are shown in the Venn diagrams (for details, see Methods). Promoters or enhancers detected by REpi-seq and confirmed by bulk ChIP-seq are indicated as percentages in the center of Venn diagrams. (E–H) Evaluation of REpi-seq reproduced reads by enhancer classification. Enhancers were classified by the relative ratio log2(H3K27ac/H3K27me3), as detailed in Supplemental Methods. H3K27ac (E), H3K27me3 (F), MED1 (G), and 5hmC (H) read counts in the classified enhancers are shown in the y-axis.











