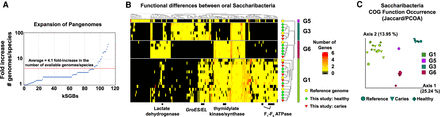
Expansion of pangenomes allows for discovery of large-scale functional differences between Saccharibacteria clades. (A) Expansion of pangenomes. Scatter plot illustrating the fold increase in the number of available genomes per species for each species in which a kSGB was recovered by this study. The red line denotes the mean of a 4.1-fold increase. (B) Differences in encoded COG functions pathways across human-associated Saccharibacteria. Heatmap illustrating the presence of various genes across four major clades of human-associated Saccharibacteria. Lactate dehydrogenase, GroES/EL, thymidylate kinase/synthase, and the F1-F0 ATPase are highlighted to illustrate differences. Rows and columns were clustered using the Jaccard distance and the “complete” clustering method. Only COG functions that were significantly different between clades are shown (adjusted Q-value < 0.05). The heatmap with all rows and columns labeled is available in Supplemental Figure S3. (C) Saccharibacteria COG function occurrence. PCoA plot illustrating a Jaccard distance matrix of COG functions across Saccharibacteria clades.











