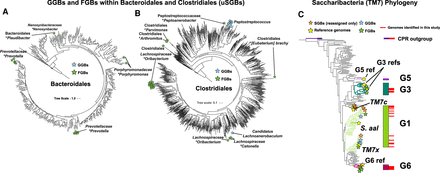
Phylogenetic trees of Bacteroidales (A), Clostridiales (B), and Saccharibacteria (C) reference genomes with placement of uSGBs. Reference genomes (C only), GGBs, FGBs, and rSGBs (C only) are denoted by stars of the indicated color. In A and B, the labels of GGBs indicate the genus of the uSGB, and the labels of FGBs indicate the family name and the name of the most closely related genus, denoted with the asterisk. In C, reference genomes and previously described oral Saccharibacteria clades G1, G3, G5, and G6 are labeled. The Bacteroidales and Clostridiales trees were constructed using PhyloPhlAn2, and the Saccharibacteria tree was constructed using Anvi'o. The trees with all leaves labeled are available in Supplemental Figure S2.











