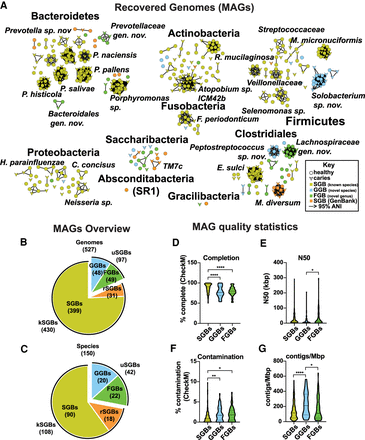
Five hundred twenty-seven metagenome-assembled genomes (MAGs) were recovered. (A) Recovery of 151 species-level genome bins (SGBs), representing 527 MAGs and 42 novel taxa. Network representing an average nucleotide identity (ANI) distance matrix, generated by fastANI (Jain et al. 2018). Nodes represent MAGs, and edges represent an ANI > 95% (the cutoff chosen to designate species boundaries in this study). Circular nodes indicate MAGs recovered from healthy samples, and chevrons indicate MAGs recovered from caries samples. Nodes are colored based on bin designation: species-level (SGB: known species; yellow), genus-level (GGB: known genus, novel species; blue), family-level (FGB: novel genus and species; green), or reassigned to SGB (rSGB: orange). Subnetworks of interest are labeled with taxonomic names. (B,C) MAGs overview. Pie charts indicating the breakdown of bin types for genomes (B) and species (C). (D–G) Statistics indicating MAG quality. Violin charts illustrating the completion (D), N50 (E), contamination (F), and contigs/Mbp (G) of the SGBs, GGBs, and FGBs. Completion and contamination were determined by CheckM (Parks et al. 2015). Statistically significant differences between groups were determined using a Tukey's multiple comparisons posttest following a one-way ANOVA: (*) P < 0.05; (**) P < 0.01; (****) P < 0.0001.











