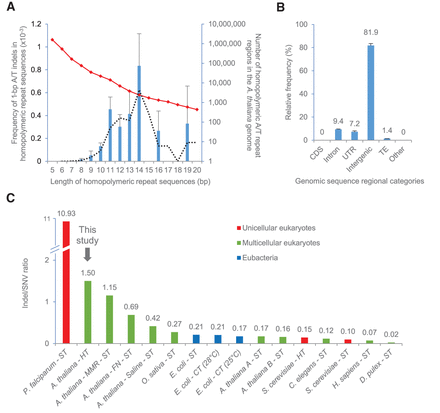
HT-induced indels cluster in homopolymeric/microsatellite stretches and cause extreme indel/SNV bias. (A) A histogram showing the frequency (left y-axis) of 1-bp A or T indels found in homopolymeric repeat regions of different lengths (x-axis: values normalized with respect to the numbers of each length category in TAIR10, numbers as indicated by the red diamond-marked line [right y-axis]) (see also Supplemental Fig. 5A,B). The black dotted line indicates a moving average trendline for indel frequency. (B) Genomic distribution of indels in HT MA lines. (CDS) Coding DNA sequence, (UTR) untranslated region, (TE) transposable element, (Other) pseudogenes and noncoding RNAs. (C) Indel/SNV ratio comparisons; data from unicellular eukaryotes (red bars), multicellular eukaryotes (green bars), and eubacteria (blue bars). Organisms were grown at relative high temperature (HT), standard temperature (ST), or alternative cold temperature (CT), as indicated. The studies shown are: Plasmodium falciparum – ST (Hamilton et al. 2017); Arabidopsis thaliana – HT (this study); A. thaliana - MMR – ST (Belfield et al. 2018); A. thaliana - FN – ST (Belfield et al. 2012); A. thaliana - Saline – ST (Jiang et al. 2014); Oryza sativa – ST (Yang et al. 2015); Escherichia coli – ST (Chu et al. 2018); E. coli – CT (28°C) (Chu et al. 2018); E. coli – CT (25°C) (Chu et al. 2018); A. thaliana A – ST (Ossowski et al. 2010); A. thaliana B – ST (Jiang et al. 2014); Saccharomyces cerevisiae – HT (Huang et al. 2018); Caenorhabditis elegans – ST (Meier et al. 2014); S. cerevisiae – ST (Liu and Zhang 2019); Homo sapiens – ST (Besenbacher et al. 2016); and Daphnia pulex – ST (Keith et al. 2016). Error bars (A,B) indicate SEM from six HT biological replicates.











