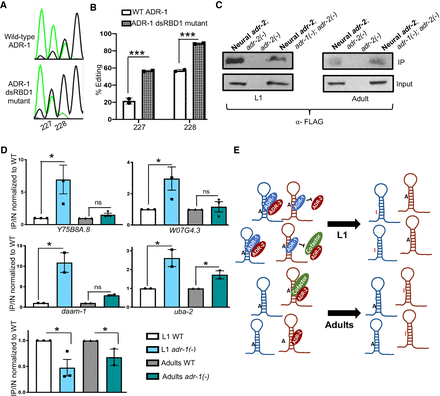
ADR-1 inhibits editing at transcripts that show increased editing in neural development by binding. (A) Sanger sequencing chromatograms of cDNA amplified from Y75B8A.8 reporter RNA isolated from the indicated worms at the L1 stage. (B) Editing in A was quantitatively measured. The average of two independent replicates is plotted; error bars, SEM. Two-way ANOVA was used to determine statistical significance. (***) P < 0.001. (C) Lysates from the indicated worm strains were incubated with FLAG magnetic beads. A portion of the lysates before immunoprecipitation and the IPs was subjected to immunoblotting with a FLAG antibody (Sigma-Aldrich F1804). (D) Bar graph represents the fold enrichment of cDNA present in the IPs divided by cDNA present in the input lysates from the indicated worm strains and normalized to the same ratio in adr-1; adr-2(-) worms. The average of three (Y75B8A.8, WO7G4.3 and lam-2) and two (daam-1 and uba-2) independent replicates were plotted; error bars, SEM. One-way ANOVA was used to determine statistical significance. (*) P < 0.05; (ns) P > 0.05. (E) Proposed model for editing regulation of neural transcripts that show increased editing during development.











