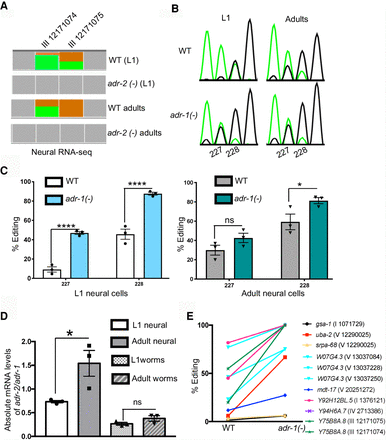
The inhibitory role of adr-1 on editing of neural transcripts is developmentally regulated. (A) Representation of neural RNA-seq reads covering the Y75B8A.8 transcript. Reads from adr-2(-) are negative controls. Green represents adenosine and brown represents guanosine at the marked chromosomal positions. (B) Sanger sequencing chromatograms of cDNA amplified from Y75B8A.8 reporter RNA. Editing sites are listed below the chromatogram. Sites 227 and 228 correspond to chromosomal positions of Chr III: 12,171,074 and 12,171,075. The nucleotides at each position are represented with a different color (green, adenosine; black, guanosine; blue, cytidine; red, thymidine). (C,D) The average of three independent replicates was plotted; error bars, SEM. One-way ANOVA was performed to determine the statistical significance. (****) P < 0.0001; (*) P < 0.05; (ns) P > 0.05. (C) Editing levels of the Y75B8A.8 neural reporter were measured in wild-type and adr-1(-) worms in L1 (left) and adult (right) stages using Sanger sequencing. (D) qPCR was performed to measure the absolute mRNA levels of adr-2 relative to adr-1 in both life stages. (E) Neural RNA-seq data were used to plot the L1 editing levels of representative sites from the group of neural RNAs that showed increased editing in development (P < 0.01, pairwise comparison using two-way ANOVA).











