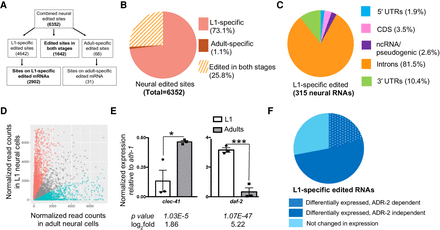
Differential expression of many transcripts during neural development contributes to the stage-specific neural editomes. (A) Workflow for identifying stage-specific edited sites following the quantification of all the neural editing sites. (B) Distribution of stage-specific edited sites. (C) Genomic distribution of L1-specific edited transcripts based on the genomic location of identified edited sites. (CDS) Coding region; (UTR) untranslated region; (ncRNA) noncoding RNA. (D) Dots represent individual genes that are down-regulated (red; 4476, log2fold < −0.5), up-regulated (blue; 5171, log2fold > 0.5), or not significantly different (gray; 7697, P-adj > 0.05) between three biological replicates of wild-type neural L1 and adult RNA-seq data. (E) The average of three independent replicates of qPCR validation of neurodevelopmentally, differentially expressed genes is plotted; error bars, SEM. Statistical significance was calculated by Student's t-test. (*)P < 0.05; (***) P < 0.001. (F) Representation of the L1-specific edited transcripts based on differential expression during neural development. Dark blue represents transcripts differentially expressed in neural development and ADR-2 dependent (dotted) or independent (solid). Light blue represents transcripts that are not differentially expressed in neural development.











