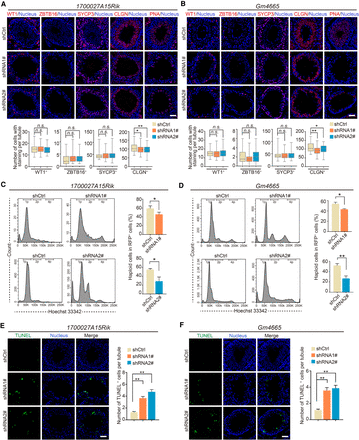
Knockdown of lncRNA candidates leading to defects in late spermatogenesis. (A,B) Immunostaining of testis sections from the control (shCtrl), 1700027A15Rik and Gm4665 knockdown (shRNA1#, shRNA2#) mice for marker proteins (red): WT1, ZBTB16, SYCP3, CLGN, PNA. Nuclei were stained with DAPI (blue). (Top) Representative images; (bottom) number of cells with staining per tubule. Data represent the mean ± SEM of at least 100 seminiferous tubules from three mice. (*) P < 0.05, (**) P < 0.01, (n.s.) P > 0.05 compared with shCtrl, Student's t-test. Scale bar, 50 μm. (C,D) Percentage of RFP+ haploid cells isolated from the control (shCtrl), 1700027A15Rik and Gm4665 knockdown (shRNA1#, shRNA2#) testes by FACS based on fluorescent labeling with Hoechst 33342. (Left) Representative flowcytometry histograms; (right) percentage of haploid cells. Data represent mean ± SEM (n = 4). (*) P < 0.05, (**) P < 0.01 compared with shCtrl, Student's t-test. (E,F) TUNEL assay of testis sections from the control (shCtrl), 1700027A15Rik and Gm4665 knockdown (shRNA1#, shRNA2#) mice. (Left) Representative images; (right) number of apoptotic cells per tubule. Data represent the mean ± SEM of at least 50 seminiferous tubules from three mice. (**) P < 0.01 compared with shCtrl, Student's t-test. Scale bar, 50 μm.











