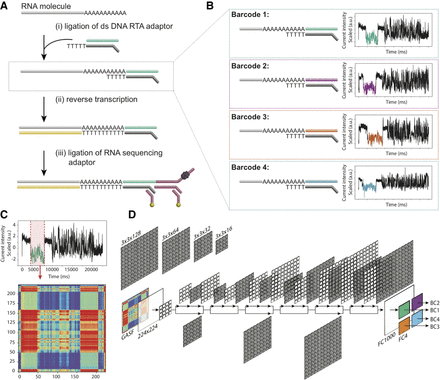
Schematic overview of the direct RNA barcoding and demultiplexing strategy. (A) Overview of Oxford Nanopore library preparation protocol for native RNA sequencing. (B) Adaptation of A to include custom DNA barcodes. (C) Barcode segmentation and transformation, where the electric current associated with a barcode adapter (highlighted in red) is extracted and converted into an image using GASF transformation. (D) Deep learning is used to classify the segmented and GASF-transformed squiggle signals into their corresponding bins, without the need of base-calling the underlying sequence. The convolution architecture of the final residual neural network classifier (ResNet-20) described in this work: FC = fully connected layer.











