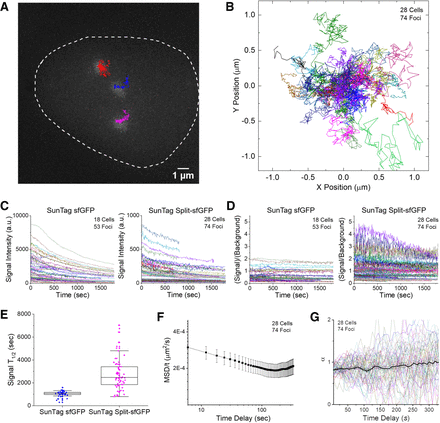
Long-term tracking of the C9-1 loci reveals diffusion behavior dependent on the observation time scale. (A) Representative trajectories of three C9-1 foci in a single nucleus, shown along with the first-frame image of GFP channel,
traced from a 2D-projected z-stack movie of live AD-293 cells integrated with the SunTag split-sfGFP CRISPR system. The trajectories
represent the loci movement for 30 min with 6-sec frame intervals. (B) Collected trajectories of C9-1 foci (n = 74). All trajectories start at the origin. (C) Traces of signal intensity of C9-1 foci shown in B (n = 74) compared with those observed in another stable AD-293 cell line expressing the SunTag sfGFP CRISPR system (n = 53). Note the 10-fold difference in scale between two graphs. (D) Corresponding traces of S/B of C9-1 foci shown in C for the SunTag sfGFP and SunTag split-sfGFP systems, with the same color codes. (E) Decay half-life of fluorescence signal of C9-1 foci with the SunTag sfGFP and SunTag split-sfGFP systems, measured by fitting
the traces in C longer than 600 sec to exponential decay functions with zero offset. (F) Mean-square displacement (MSD) divided by time, calculated over varying time delays. Error bars represent SEM between observed
foci. (G) Diffusion exponent α found by fitting MSD curves of individual trajectories to 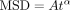 locally at varying time delays. Average and SEM are shown as a black curve.
locally at varying time delays. Average and SEM are shown as a black curve.











