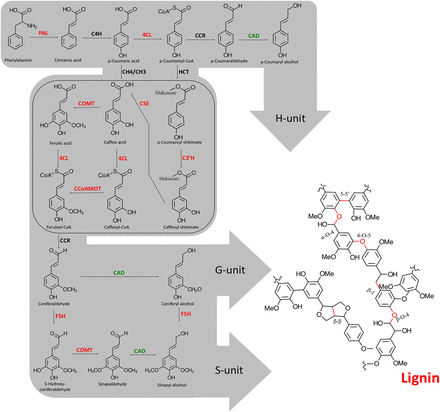
Summary of the lignin biosynthetic pathway in higher plants. Enzymes are abbreviated as follows: (PAL) phenylalanine ammonia-lyase; (C4H) cinnamate 4-hydroxylase; (4CL) 4-coumarate:CoA ligase; (C3H) p-coumarate 3-hydroxylase; (C3′H) p-coumarate shikimate 3-hydroxylase; (HCT) shikimate/quinate hydroxycinnamoyl transferase; (CCR) cinnamoyl-CoA reductase; (CAD) cinnamyl alcohol dehydrogenase; (CSE) caffeoyl shikimate esterase; (COMT) caffeic acid O-methyltransferase; (CCoAOMT) caffeoyl-CoA O-methyltransferase; (F5H) ferulate 5-hydroxylase. Enzymes in red were found to be directly connected to MYB125 in the gene expression network and to be differentially expressed when MYB125 is overexpressed in poplar transgenic roots. The enzyme in green was not directly connected to MYB125 in the gene expression network, but single regression analysis showed significant association with the local-regulatory SNPs of MYB125. The box includes the most recent step discovered in the lignin biosynthesis pathway in plants.











