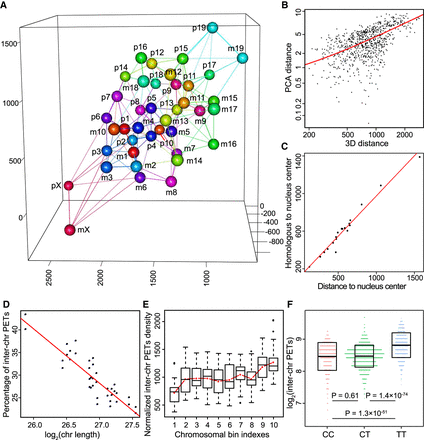
Spatial organization of chromosomes in the 3D nucleus and principles of nucleus organization. (A) Constructed 3D nucleus at chromosomal resolution. Each chromosome and its five closest chromosomes are connected by lines: (p) paternal chromosomes; (m) maternal chromosomes. (B) Chromosomal interaction pattern is positively correlated with chromosomal distance in the 3D nucleus (R = 0.67, P-value < 2.2 × 10−16). (C) Distances to the center of the 3D nucleus of paternal chromosomes are highly correlated with that of their maternal homologous counterparts. Center of the 3D nucleus is calculated as the mean coordinate of all chromosomes (R = 0.98, P-value = 1.2 × 10−14). (D) The lengths of chromosomes are negatively correlated with the percentages of inter-chromosomal interactions (R = 0.9, P-value = 1.5 × 10−14). (E) Inter-chromosomal interactions are increasing from centromere to telomere along the chromosomes. Each chromosome is evenly divided into 10 bins. The red line plots the mean value for each bin. (F) Telomere ends have much stronger inter-chromosomal interactions than that of centromere ends. Each chromosome is evenly binned into three parts, namely centromere end (C), middle (ignored), and telomere end (T). P-values were calculated using unpaired t-test.











