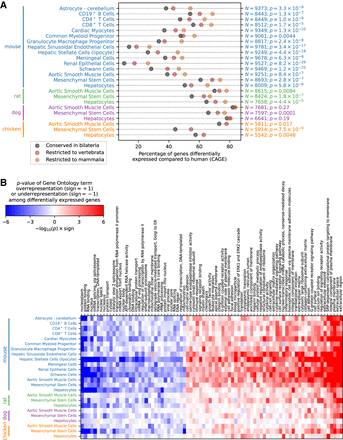
Conservation and Gene Ontology analysis of differentially expressed genes. (A) Percentage of differentially expressed genes in each cell type as a function of age of the most recent common ancestor. The one-sided P-value of a Poisson regression model against the evolutionary age category is shown on the right, together with the number N of expressed genes in each cell type with an annotation in the NCBI HomoloGene database. (B) Gene Ontology analysis of differentially expressed genes. The P-value, calculated using Fisher's exact test, of overrepresentation or underrepresentation of differentially expressed genes in each Gene Ontology category compared to an expression-matched set of background genes is shown in red and blue, respectively.











