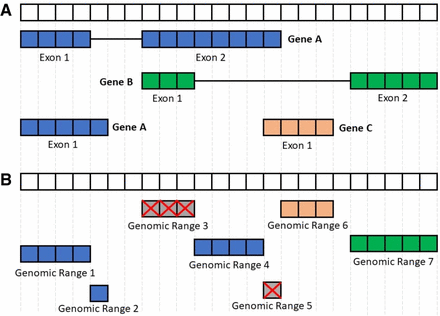
Processing the FANTOM-CAT genomic ranges. This figure summarizes the disjoining and exon disambiguation processes performed before extracting expression information from recount2 using the FANTOM-CAT gene models. (A) Representation of a genomic segment containing three distinct, hypothetical genes: gene A having two isoforms, and genes B and C with one isoform each. Each box can be interpreted as one nucleotide along the genome. Colors indicate the three different genes. (B) Representation of disjoint exon ranges from example in panel A. Each feature is reduced to a set of nonoverlapping genomic ranges. The disjoint genomic ranges mapping back to two or more distinct genes are removed (crossed gray boxes). After removal of ambiguous ranges, the expression information for the remaining ones is extracted from recount2 and summarized at the gene level.











