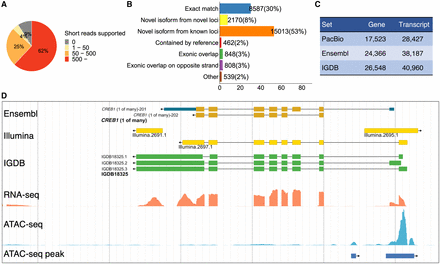
Gene model construction. (A) Splice junctions from the PacBio set validated by the Illumina short reads. Splice junctions are classified into four groups depending on the numbers of short reads supported. (B) Comparison of the PacBio set with the Ensembl set. The largest class is “novel isoforms from known loci.” (C) Numbers of genes and transcripts in three model sets. (D) An example showing the improved annotation of the medaka genome. The Ensembl model is correct overall. The Illumina models missed isoforms or are likely inaccurate owing to transcriptional noise. The IGDB model identified multiple novel isoforms with accurate boundaries. The ATAC-seq clearly showed the activity of the promoter.











