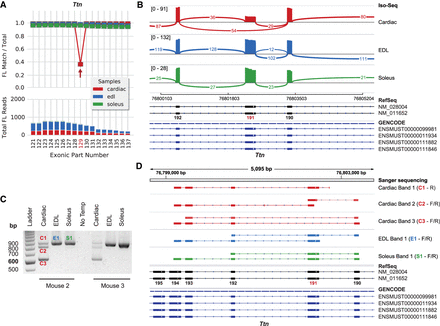
Titin exon 191, an undocumented cassette exon removed from a subset of cardiac transcripts. (A) Titin exon coverage graph (cropped) produced by exCOVator. (Top) Line graph; the red arrow points to exonic part (EP) 129 or exon 191 of NM_011652 (N2-A). (Bottom) Stacked bar graph displays full-length (FL) read coverage of EP. (B) Sashimi plot showing the same data in the Integrative Genomics Viewer (IGV). The plot shows consensus reads from the BAM file (not FL reads). Minimum splice junction coverage = 5. (C) Agarose gel of RT-PCR products from soleus, EDL, and heart. Primers target exons 189 and 194 with predicted product sizes of 882 bp (+exon 191, C1) and 615 bp (no exon 191, C3). All soleus and EDL transcripts include exon 191. Cardiac has both predicted bands and one unknown band in the middle (C2) that is likely a heteroduplex. (D) Sanger sequencing data of products from C displayed using the IGV BLAT tool. Exon 191 is missing only from cardiac band 3 (C3) and is present in all other tissues.











