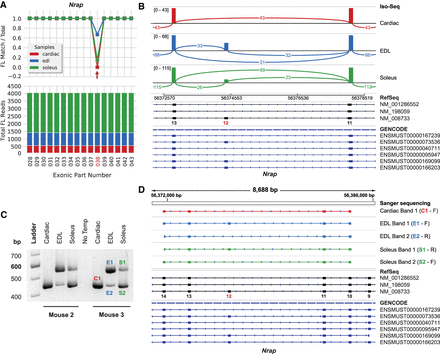
Differential usage of Nrap exon 12 (exonic part 038). (A) Nrap exon coverage graph (cropped) produced by exCOVator. (Bottom) Stacked bar graph showing total full-length (FL) read coverage of all exonic parts (EP) for the gene. (Top) Line graph displaying the ratio of [FL reads matching the EP/total FL reads] that overlap the EP coordinates. (B) Sashimi plot from the Integrative Genomics Viewer (IGV) displaying differential splicing of exon 12. The plot displays consensus reads from the BAM file (not FL reads). Minimum splice junction coverage = 5. (C) Agarose gel showing RT-PCR products pertaining to exon 12 from soleus, EDL, and heart. Primers target exons 9–14 (549 bp includes and 444 bp excludes exon 12). (D) Sanger sequencing of products excised from the gel in C. The top half shows sequences aligned using the IGV BLAT tool. Cardiac band 1 and EDL and soleus band 2 are missing exon 12.











