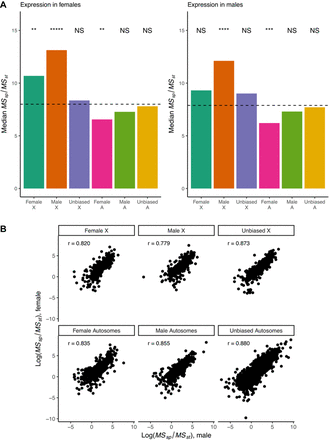
Ratio of interspecific to intraspecific variability in expression for genes in the Drosophila brain. (A) Male-biased genes on the X Chromosome show significantly elevated variability ratios, consistent with directional selection. Similarly, female-biased genes on the X Chromosome show signatures of directional selection in females. Female-biased autosomal genes show significantly reduced ratios, consistent with balancing selection on gene expression. Significance of medians relative to the genome-wide median is shown. P-values determined using 1 million simulations per group and Benjamini–Hochberg correction. (*****) P-value <1 × 10−5; (****) P-value <1 × 10−4; (***) P-value <1 × 10−3; (**) P-value <1 × 10−2; (*) P-value <5 × 10−2. A boxplot version of the data showing dispersion around the median is presented in Supplemental Figure S4. (B) Ratios of interspecific to intraspecific variability in males (on x-axis) show large correlations (Spearman's) with ratios in females (y-axis) irrespective of sex bias or chromosomal location. Only 1:1:1 orthologs were considered. Genes were considered male- or female-biased if they showed that bias in at least one species at an FDR of 0.05. For any given gene, the chromosomal location was taken to be the location of the D. melanogaster ortholog.











