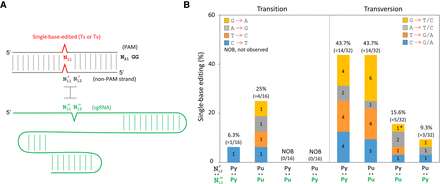
Genome-wide single-base editing aided by target-mismatched sgRNAs in CRISPR-Cas9. (A) Design of mismatched sgRNAs for single-base-edited targets. Single mismatches (N′12 : N′′12) between non-PAM strands and sgRNAs were introduced immediately after the single-base-edited sites (N11). Note that mismatches in two galK ([504] and [578]) targets were designed at N′8 : N′′8 and N′17 : N′′17, respectively. (B) Single-base genome editing for 16 different DNA targets was aided by various base-parings (N′12 : N′′12). Numbers in parentheses and colored boxes are the frequency and the successful events of single-base editing, respectively. Asterisk indicates one single-base editing was observed using a target-matched sgRNA among 16 target-matched sgRNAs.











