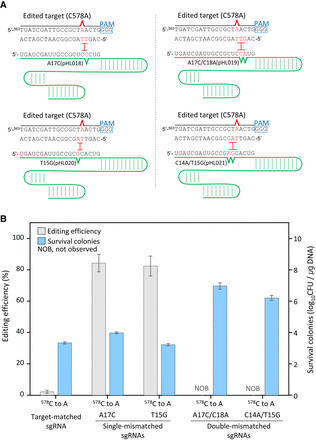
Single-base editing in the PAM-proximal region using target-mismatched sgRNAs. (A) Negative selection of edited target sequence (C578A in the galK gene) by Cas9/target-mismatched sgRNA complexes. Single or double mismatch(es) were located adjacent to the C578A point mutation site. (B) Genome editing efficiencies using target-matched, single- or double-base mismatched sgRNAs and the number of surviving cells. Each bar represents the mean of three independent experiments.











