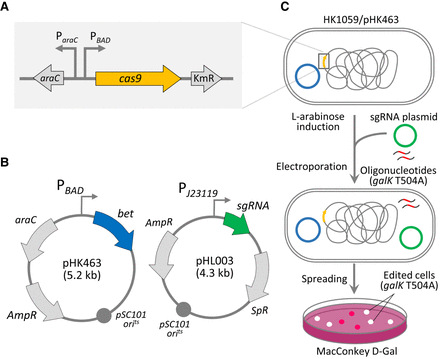
Schematic representation of bacterial genome-editing. (A) Chromosomal construction of cas9 downstream from the PBAD promoter, which forms an L-arabinose-inducible cas9 gene. (B) Construction of plasmids. Lambda bet expression vector (pHK463) was derived from pKD46 plasmid. The sgRNA expression plasmid (pHL003) harbors a temperature-sensitive origin for iterative genome engineering. (C) Genome editing. Mutagenic oligonucleotides carrying galK T504A and sgRNA plasmids were electroporated into E. coli cells overexpressing Cas9 and Bet proteins by L-arabinose induction. Recovered cells were spread on MacConkey agar containing D-galactose. Red/white colonies were counted for determination of galK editing efficiency.











