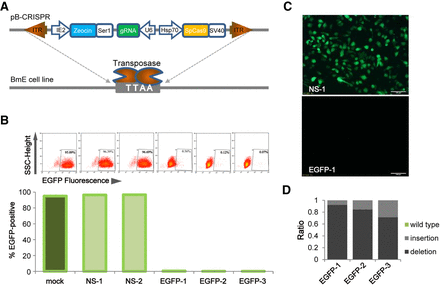
The piggyBac library delivery method for gene knockout in B. mori cells. (A) Schematic of the piggyBac library delivery vector (pB-CRISPR): (ITR) inverted terminal repeats of piggyBac; (IE2) IE2 promoter; (Zeocin) zeocin selection marker gene; (Ser1) Sericin 1 poly(A); (gRNA) sgRNA and Scaffolds; (U6) polymerase III U6- promoter; (Hsp70) hsp70 promoter; (SpCas9) B. mori codon-optimized SpCas9 coding sequence; (SV40) SV40 poly(A). (B) Flow cytometry analysis (upper) and statistical analysis (lower) of BmE-Mi-EGFP cells transduced with pB-CRISPRs: (mock) no vector; (NS-1) pB-CRISPR-NS-1; (NS-2) pB-CRISPR-NS-2; (EGFP-1) pB-CRISPR- EGFP-1; (EGFP-2) pB-CRISPR-EGFP-2; (EGFP-3) pB-CRISPR-EGFP-3. (C) Fluorescent images of BmE-Mi-EGFP knockout experiments. (D) Sanger sequence analysis of the ratio of indels for EGFP-1 (left), EGFP-2 (middle), and EGFP-3 (right) target regions of BmE-Mi-EGFP cells.











