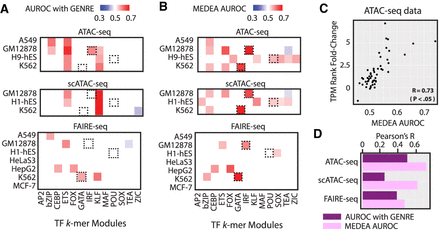
Generalization of MEDEA to other types of chromatin accessibility assays. (A,B) For the peaks obtained with the indicated chromatin accessibility assays in indicated cell lines, the AUROC enrichment of the 13 benchmarking motifs evaluated by using either (A) AUROC with GENRE background or (B) MEDEA. MEDEA-filtered peaks were obtained by contrasting the peaks in the indicated assays with the default panel of 12 ENCODE-DREAM DNase-seq data sets. Black boxes as in Figure 1A and Figure 2, B through D. (C) For the bulk ATAC-seq data sets, scatterplot to correlate the MEDEA AUROC (top panel in B) with the associated transcriptomic up-regulation (y-axis values from Fig. 1B). (D) Barplot to compare the correlation coefficients (Pearson's R) between TF up-regulation and motif enrichment obtained with the different assays, either by using (1) AUROC with GENRE background (purple bars) or (2) MEDEA AUROC (pink bars). For additional scatterplots depicting correlations for other assay/method combinations, see also Supplemental Figure S15.











