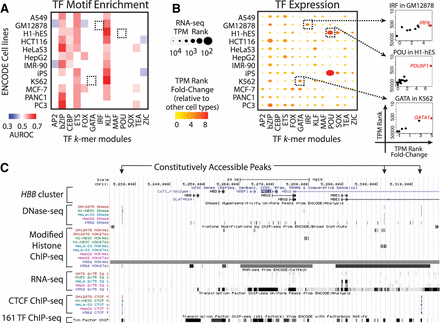
TF motif enrichment in DNase-seq peaks partially infers lineage specifiers. (A) For a benchmarking set of 13 TF binding motifs, AUROC enrichment in a panel of 12 ENCODE cell lines was calculated using the Glossary motif enrichment tool with background sequences generated with GENRE software. Black boxes highlight three motifs (IRF, POU, GATA) expected to be enriched in the corresponding cell lines (GM12878, H1-hES, K562, respectively). (B) For each indicated cell type and TF family, TPM rank (y-axis) and TPM rank fold-change (x-axis) of the most up-regulated TF. Each TF's rank is determined according to RNA-seq TPM levels of all genes and serves as a measure of the TF's expression. The TPM rank fold-change of each TF is assessed by comparing the TF's rank in the indicated cell type to the TF's median rank across the remaining cell types as a measure of the TF's up-regulation. (Right insets) TPM rank and TPM rank fold-change evaluation of the TFs in the indicated families and cell types. The values and names of the most up-regulated TFs are colored in red. (C) Screenshot of the UCSC Genome Browser for the hemoglobin subunit beta (HBB) cluster locus (Chr 11: 5,220,000–5,320,000). Genomic profiles of five cell lines used in this study are from ENCODE. Three locations of constitutively accessible peaks are highlighted.











