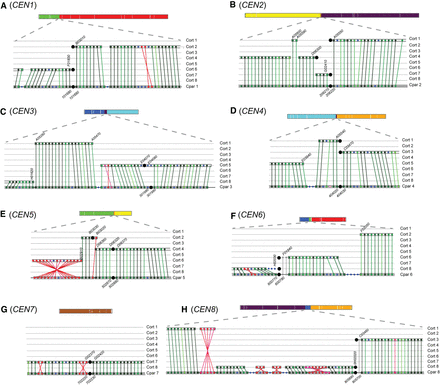
Interspecies synteny breakpoints occur at centromeres. Synteny between C. parapsilosis and C. orthopsilosis was visualized using SynChro (Drillon et al. 2014), with a delta value of two. Changing delta values had minor effects on predicted synteny. A diagrammatic representation of each C. parapsilosis chromosome, colored as in Figure 4C, is shown to scale at the top of each panel. The lower sections of each panel show the gene order around the centromere. (A–H) Gene order around the eight centromeres in C. parapsilosis compared with C. orthopsilosis. The bottom row in each panel shows gene order on the C. parapsilosis chromosome, and the eight C. orthopsilosis chromosomes are shown above. Each gene is indicated by a colored dot, and RBHs are joined by lines. Syntenic blocks are surrounded with a box. Centromeres are shown by large black circles. The chromosome number is indicated at the side of each panel. The names of some genes are shown for orientation purposes. We removed the prefix “CORT0” from C. orthopsilosis genes and “CPAR2_” from C. parapsilosis genes for brevity. The color of the dots indicates the similarity of the proteins. Noninverted RBHs are shown in green, ranging from darkest (>90% similarity) to lightest (<30% similarity), and inverted orthologs are shown in red. Genes without RBH orthologs are shown in blue. Genes in gray were not identified as RBHs by SynChro but were identified using CGOB (Fitzpatrick et al. 2010; Maguire et al. 2013).











