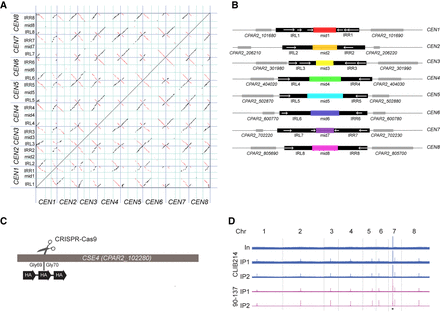
C. parapsilosis centromeres consist of unique mid regions surrounded by partially conserved inverted repeats (IRs). (A) Dot matrix plot comparing the putative centromere sequences in C. parapsilosis. Centromere regions (see Supplemental Table S2) were concatenated and are delineated by dark blue lines. IRs (right, IRR; left, IRL) are separated with cyan lines. Each dot represents a 25-bp window. Inverted sequences are shown in red; direct repeats, in black. (B) Diagrammatic representation of the information in A. Regions that are conserved among chromosomes are shown in black. Locations of IRs (>75% DNA sequence identity) are shown with white arrows. The mid regions are illustrated in different colors that indicate that each of them has a unique sequence. Adjacent genes are shown in gray. Each region shown is ∼10 kb in length. (C) Three copies of an HA tag were introduced into both alleles of the endogenous CSE4 gene in C. parapsilosis CLIB214 and 90-137 using CRISPR-Cas9 editing. The gene was cut between glycine 69 and glycine 70, and a repair template containing the HA tags was inserted by homologous recombination. The construct was confirmed by sequencing. (D) Visualization of the ChIP-seq signal across all chromosomes (Chr) in Cse4-tagged derivatives of C. parapsilosis CLIB214 and 90-137. (In) Input (before immunoprecipitation); IP1 and IP2 show two independent immunoprecipitation replicates from each strain. Strains derived from C. parapsilosis CLIB214 are shown in blue; from 90-137, in purple. There is one signal per chromosome in the IP samples, identifying the centromere, except for Chromosome 7, in which the rDNA locus (black asterisk) also generates a signal. The x-axis in each plot is the chromosome coordinates, and the y-axis is the number of reads mapping to a position. The maximum scale for C. parapsilosis CLIB214 is restricted to reduce the signal from the rDNA. Data are visualized using Integrative Genomics Viewer (IGV) (Thorvaldsdóttir et al. 2013).











