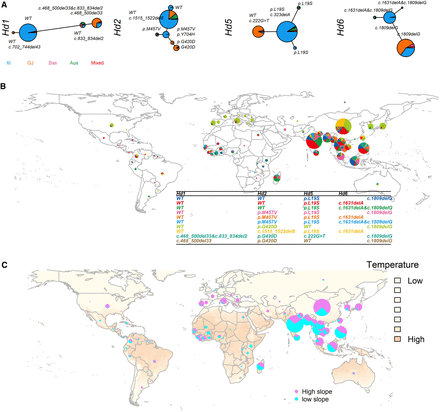
Natural allelic variations in the Hd1, Hd2, Hd5, and Hd6 genes among 3000 rice genome accessions. (A) Haplotype network of four genes from 3010 rice accessions. Haplotype frequencies are proportional to the sizes of the circles. Each major haplotype is annotated with the functional sites. The total number of accessions used for haplotype analysis varies among four genes. The different colors represent the classification of rice accessions. (B) Geographic distribution of the four-gene combinations in rice accessions. The different colors represent different combinations of Hd1, Hd2, Hd5, and Hd6. The relative size of each pie indicates the ratio of accessions within each combination in a given country. (C) Geographic distribution of the multigene haplotypes with different slope values indicating the sensitivity to temperature. High slope, absolute value ≥ 2; low slope, absolute value < 2.











