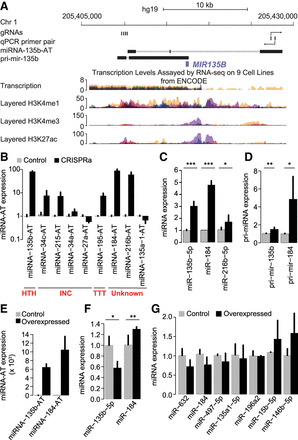
The act of antisense transcription, rather than the transcripts themselves, regulates the expression of sense miRNAs. (A) The various genomic and chromatin features of a sense–antisense miRNA pair were shown from top to bottom: (1) the positions of the four gRNAs used for miRNA-135b-AT activation; (2) the location of the primer pair used to measure miRNA-135b-AT expression; (3) the annotation of the full-length miRNA-135b-AT from FANTOM5; (4) the annotation of the pri-mir-135b transcript from previous studies; (5) transcription levels assayed by RNA-seq on nine cell lines from ENCODE; (6) three histone marks associated with regulatory elements and promoter from the ENCODE. (B–D) RNA expression of miRNA-ATs (B), sense-mature miRNAs (C), and sense pri-miRNAs (D) in HEK293T cells transfected simultaneously with four gRNAs targeting the indicated miRNA-ATs along with the dCas9-activator construct. Control cells were transfected with the indicated guide RNAs alone. (Error bars) SD based on three biological replicates. P-values were calculated using student's t-test. (E,F) RNA expression of miRNA-ATs (E) and sense-mature miRNAs (F) in HEK293T cells transfected with expression vectors harboring individual miRNA-ATs. Control cells were transfected with the empty expression vectors. (Error bars) SD based on three biological replicates. P-values were calculated using student's t-test. The expression level of miR-135b-5p in F was quantified using Qsep1 but not real-time PCR (Supplemental Fig. S13). (G) RNA expression of sense-mature miRNAs in HEK293T cells transfected with expression vectors harboring individual miRNA-ATs. Control cells were transfected with the empty expression vectors. (Error bars) SD based on three biological replicates. P-values were calculated using student's t-test, and no significant changes were observed between overexpressing and control cells for all miRNAs studied.











