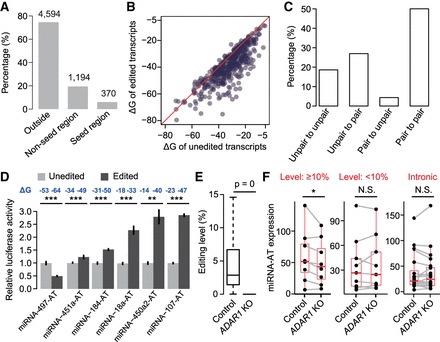
RNA editing modulates miRNA-mediated regulation of miRNA-ATs. (A) The percentage of miRNA-AT editing sites that occur in the positions opposite to the sense miRNAs. (Outside) Outside of the region opposite to the mature miRNA. The number of editing sites is indicated above the bar. miRNA-ATs with their intronic regions targeted by the paired sense-mature miRNAs were excluded. (B) Comparison of the free energy (ΔG) between unedited and edited regions of miRNA-ATs that are opposite to the paired pre-miRNAs. For a miRNA-AT with multiple editing sites, a fully edited form was considered. miRNA-ATs with their intronic regions targeted by the paired sense-mature miRNAs were excluded. (C) The proportion of editing sites with different types of pairing patterns before and after editing. For each editing site, we predicted the secondary structure of the miRNA-ATs before and after editing and compared the pairing patterns of this site. (D) Dual-luciferase reporter assays to compare the effect of sense miRNAs on unedited and edited forms of miRNA-ATs. For each miRNA-AT, the predicted RNA secondary structures of unedited and edited versions were shown in Supplemental Figure S11. (Relative activity) Average Renilla luciferase activity normalized to firefly luciferase in four biological replicates; (error bars) SD based on four biological replicates. P-values were calculated using student's t-test. (E) Editing level comparison between control and ADAR1 knockout HEK293 cells. All known human editing sites obtained from RADAR database version 2 were used for analysis. Sites that were edited in control sample and covered by at least 30 reads in both samples were selected. P-values were calculated using paired Wilcoxon signed-rank test. (F) Comparison of miRNA-AT expression levels between control and ADAR1 knockout HEK293 cells. We required that miRNAs were expressed in HEK293 cells. P-values were calculated using the paired Wilcoxon signed-rank test. (Level ≥ 10%) The regions of miRNA-ATs that are opposite to pre-miRNAs are located in the exonic regions and have at least one site with editing level ≥ 10%; (level < 10%) control set 1, the regions of miRNA-ATs that are opposite to pre-miRNAs are located in the exonic regions and have all sites with editing level < 10%; (intronic) control set 2, the regions of miRNA-ATs that are opposite to pre-miRNAs are located in the intronic regions.











