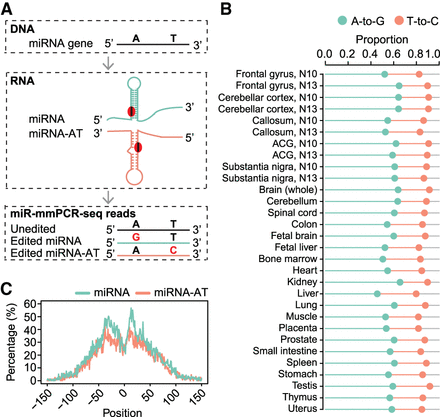
Extensive editing of miRNA-ATs. (A) Schematic of miRNA-AT identification via A-to-I RNA editing detection using miR-mmPCR-seq. (B) The percentage of A-to-G and T-to-C mismatches among all 12 mismatch types in miRNA loci in different samples. Sites with editing level ≥2% were used for analysis. Orange and green lines represent the proportions of T-to-C and A-to-G variants, respectively. (C) Metagene profiles depicting the editing site distribution surrounding pre-miRNAs and miRNA-ATs. Position 0 of a miRNA is defined as the central position of the loop region of a pre-miRNA. Position 0 of a miRNA-AT is defined as the position opposite to the central position of the loop region of a pre-miRNA. The negative value is the distance between an upstream position and the central position, and the positive value is the distance between a downstream position and the central position. Percentage of As that were edited at each position was calculated. Sites with editing level ≥2% were used for analysis.











