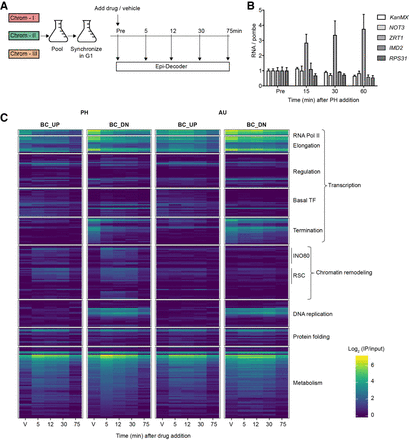
Chromatin-proteome dynamics at the Barcoded-HO locus upon chemical inhibition of transcription. (A) Three versions of the chromatin TAP-tag subset, each containing a unique set of BC pairs, were combined into one pool (Chrom-3×BC). The culture was incubated with a-factor pheromone to synchronize the cells in G1, after which PH or AU was added and samples were collected at the time points indicated for Epi-Decoder analysis, RNA analysis, and flow cytometry. A sample treated with vehicle was collected at the last time point, and a control sample was taken of a G1-arrested culture without treatment. All experiments were performed at 16°C to facilitate capturing dynamic binding events. (B) Analysis of mRNA changes over time by RT-qPCR confirmed the decay of most transcripts and an increase in the PH-responsive ZRT1 gene under conditions shown in panel A. RNA levels are relative to untreated (Pre) and normalized to a transcript from a spike-in of untreated Schizosaccharomyces pombe cells (see Methods) to correct for global changes (mean of three biological replicates ± SD). (C) Heatmap of the binding scores (mean IP/input of three biological replicates) of selected proteins with a binding score >0.5 at any of the four local proteome time series indicated. For mean binding scores of all proteins examined in the Chrom-3×BC library, see Supplemental Table S2. Proteins were manually clustered and ranked in functional subcategories.











