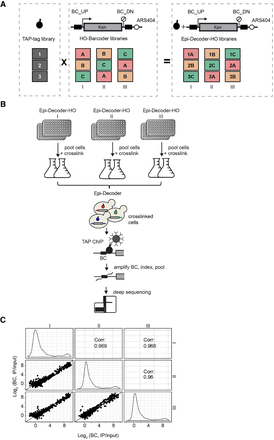
Outline of expanded and optimized Epi-Decoder analysis. (A) Three Epi-Decoder-HO libraries were generated by crossing the TAP-tag library to an expanded HO-BC library in three different ways to combine each TAP-tag protein with three independent barcodes (BCs) (see also Supplemental Fig. S1A). The HO-BC locus consists of a constitutively expressed 1.5-kb KanMX resistance gene integrated at the HO locus, controlled by the heterologous AgTEF1 promoter and terminator from Ashbya gossypii, and flanked by a promoter-proximal BC_UP and a terminator-proximal BC_DN. Downstream from BC_DN lies an origin of replication (ARS404). (B) Clones of each Epi-Decoder-HO library are combined and processed in two separate pools and used for ChIP of TAP-tagged proteins (spheres with black handle). The BCs (colored lines), which flank the KanMX reporter gene (gray box) at the HO locus, are amplified from ChIP and input and indexed, allowing for the pools to be combined and counted by massive parallel sequencing. The relative BC count (IP/input) reports on protein abundance of each TAP-tagged protein (approximately 4250) at the barcoded locus. (C) Comparison of the binding scores (IP/input) of both BC_UP and BC_DN of chromatin binders (as determined previously by Korthout et al. 2018) in the three Epi-Decoder-HO libraries. Indicated are the Spearman’s correlation coefficients, and the diagonal line represents x = y. Density plots show the distribution of the BC counts in each of the three replicates. For counts of all proteins examined, see Supplemental Table S1. The results for BC_UP and BC_DN separately are shown in Supplemental Figure S1, B and C.











