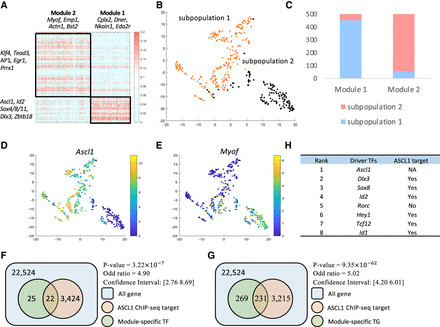
TimeReg analysis on direct reprogramming from fibroblast to neuron. (A) Heatmap of reordered normalized TRS scores at day 2. The black line represents the detected modules from NMF. (B) t-SNE plot of single-cell RNA-seq data. Color represents clustering label. (C) Distribution of day 2 top 500 module-specific genes’ maximum-expressed subpopulations from single-cell RNA-seq data. (D,E) Expression of module-specific genes on t-SNE plot. (F,G) Module 1–specific genes are enriched in ASCL1 ChIP-seq target genes. (H) List of driver regulators of Module 1 in day 2.











