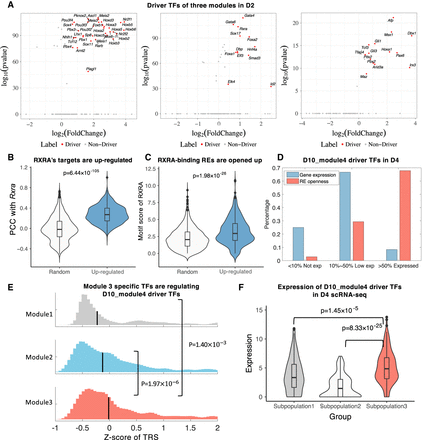
Identification of driver regulators reveals ancestor–descendant fates for regulatory modules. (A) Driver regulators of D2_Module1, D2_Module2, and D2_Module3. The x-axis is log2 fold change; the y-axis is −log10(P-value). (B) Distribution comparison of Rxra’s PCC with up-regulated genes and randomly selected genes. (C) Distribution comparison of RXRA’s motif enrichment on REs of up-regulated genes and randomly selected genes. (D) Distribution of D10_Module4 driver TF expression (blue bar) and their RE openness (red bar) on D4. Expression or openness greater than the median are labeled as “Expressed,” less than decile are labeled as “Not exp,” and the remaining are labeled as “Low exp.” (E) Distribution of Z-score of TRS between top 50 module-specific TFs and driver TFs of D10_Module4. (F) Expression distribution of the Module4's driver regulators on D4 scRNA-seq data. Columns represent subpopulations identified from scRNA-seq data.











